Persistence on the Internet: A Global Problem
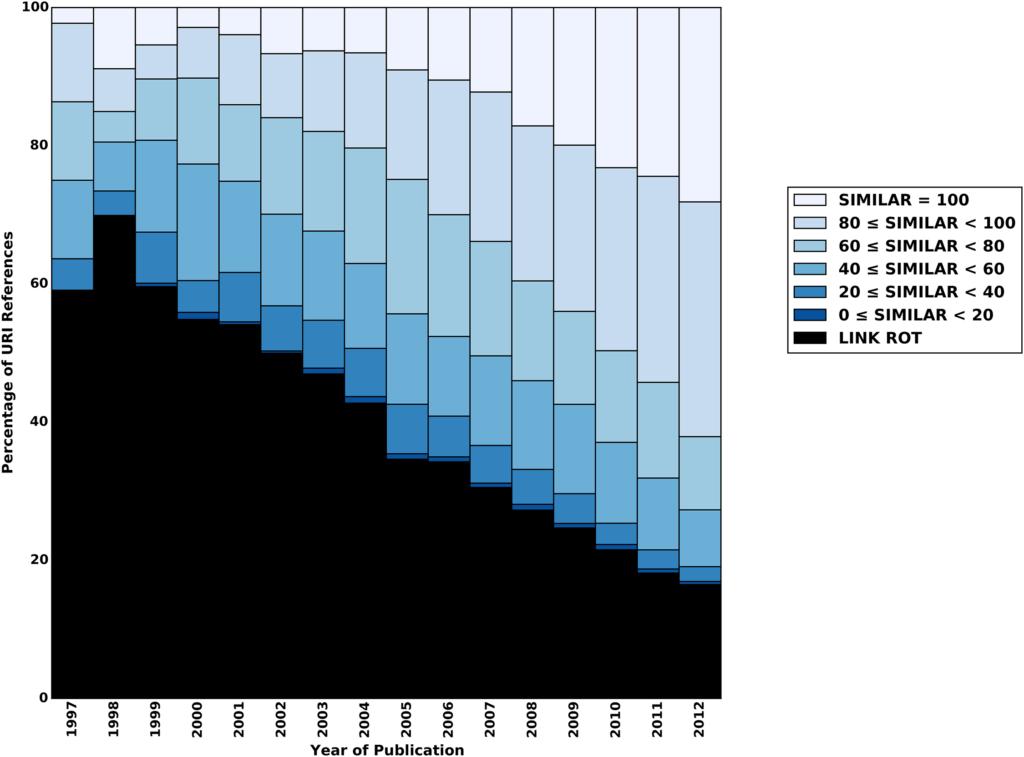
We all agree that we need a digital version of the scientific record. The problem is that the current Internet was not designed for this purpose. Beyond the issues of broken links, there is the problem of content drift, where the URL resolves but is no longer linked to the initial content. A study by Jones et al. (2016) shows that link rot or content drift affects almost all Internet references in the corpus of Elsevier, arXiv, and PubMed Central. The older the reference, the more severe the problem gets. URLs from 1997 are hardly ever valid nowadays (see Figure below). But even 50% of URI references just three years old are affected by link rot and content drift.
In this post, we discuss current approaches to addressing the shortcomings of the existing Internet technology, identify remaining bottlenecks, and suggest how they could be resolved. The upgrades to the backbone of the scientific record we discuss here would go a long way toward addressing the replication crisis and the increasing challenges for publishers to spot fake research.
Persistent Identifiers: Challenges for DOIs
Of course, persistence problems on the internet are well known. The solution is persistent identifiers (PIDs) for digital content, which are supposed to enable long-lasting references and access. The Digital Object Identifier (DOI) system was developed as the PID solution for the digital version of the scholarly record — and it’s a huge success story.
DOIs have emerged as the de-facto standard of persistent identifiers for scholarly information, up to the point where some people think DOIs are guarantees for legitimate science (they are not). Over 100 million DOIs have been registered for publications, pre-prints, scientific datasets, and other academic content. DOIs are crucial for academic databases and indexing services such as the Web of Science, Scopus, Dimensions, etc.
However, our current gold standard for identifying and resolving scholarly content is still falling short of the desired level of persistence. A truly persistent identifier should always resolve to the same result. But that’s not the case for DOIs. According to a study by Klein and Balakireva (2020), approximately 50% of DOI requests fail to resolve to their target resource. The study details how DOI behavior is inconsistent across different networks: You may get very different responses when trying to resolve a DOI from your computer at work and your mobile phone when traveling.
Obtaining DOIs and updating the DOI record is also a tedious and expensive task for publishers. Both CrossRef and DataCite have fee structures that charge more money if more DOIs are created. If the URL of a publication changes, the DOI publisher is responsible for updating the DOI database — and not all of them do so consistently and quickly. In other words, the DOI system relies on social contracts and trust between platform operators and PID providers that are difficult and expensive to enforce. This limits both the reliability and the scalability of the DOI system.
Moreover, in a potential future world of FAIR (Findable, Accessible, Interoperable, and Reusable) science, every research artifact – not just the published manuscript – will need a globally unique, persistent, and resolvable identifier. Trillions of PIDs must be minted over the next decade to achieve that goal, and trillions of PID-to-URL mappings will need to be maintained. This is inconceivable with the current DOI system.
The prevalence of link rot, content drift, artifact fragmentation, and inconsistent resolution is poised to expand alongside this explosion of (or worse, lack of) DOI minting. The social costs of not embracing FAIR science are even worse: A study on behalf of the European Commission found that the annual cost of not having FAIR research data to the European economy alone is more than €10 billion every year. This estimate does not even include the adverse effects on research quality, technological progress, and slowed economic growth.
dPID: Distributed Architectures for Distributed Ecosystems
Technological solutions to these problems have already been developed, but have yet to be widely implemented within the scientific ecosystem. Over the past two decades, several open-source communities and W3C working groups have addressed the root causes underlying the lack of persistence on the Internet. The core idea is that PIDs should not ask, “What is the content stored at this location?” Instead, the right question is, “What is the content with this digital fingerprint?” A minor tweak to such a foundational question has far-reaching impacts, enabling deterministic resolution.
Deterministic resolution is the idea that PIDs should be guaranteed to resolve to their indexed resource. Deterministic resolution has long been an elusive goal, but it is finally in sight due to the maturity of enabling technologies. This new class of PID technology is called “dPIDs” – short for “decentralized persistent identifiers.” Beyond deterministic resolution, dPIDs offer many surprising and beneficial properties that could substantially improve how science is done, communicated, and evaluated.
The act of minting a dPID enables making the underlying content available on an open peer-to-peer network where repositories, libraries, universities, and publishers can participate in curating and validating content. It also makes it possible to store redundant copies with the same identifier on different servers operated by various entities without developing or using API-based services. It also eliminates the need to maintain and update DOI records manually. dPIDs also provide a graceful way to handle the data transition to a new host when a repository or journal is scheduled to go offline or changes owners because the link to the content does not change, thus adding a built-in way to safeguard content that is otherwise at risk of being lost forever.
dPIDs are not just identifiers for a single file. Instead, they allow addressing linked folder structures that scale nearly indefinitely. Each file in this folder structure can be uniquely addressed from the base dPID, enabling digital research objects that link all relevant project pieces together (e.g., manuscripts, data, code). dPIDs are also built to be versionable, meaning that indexed content can change over time, without overwriting the original version. Changes are logged, timestamped, and digitally signed by the PID owner, providing traceable and verifiable provenance for any modifications performed to a PID, while ensuring that previous versions of the content remain equally resolvable.
Versionability and provenance of dPIDs enable researchers to create a transparent track record of how they arrived at their final results, which would be aligned with best open science practice and help to address the replication crisis and the flood of fake research that poses substantial challenges for publishers. Instead of just seeing the submitted version of a manuscript, editors, referees, and readers could also see the entire history leading up to that manuscript, including time-stamped versions of analysis plans, data, code, lab notes, early drafts, etc.
Persistent resolution also enables scientists to retrieve open datasets directly via dPID into their programming and computing environment with a single line of code or, alternatively, to send a containerized compute job to the servers that host the data. This latter technology is called edge-computing and is especially valuable for sensitive datasets that cannot be publicly shared or for extremely large datasets: For example, downloading 1PB (e.g., climate modeling data) can cost over $100,000 in egress fees and take several months even under optimal conditions – a prohibitive burden even for very well-funded scientists.
While technically simple to implement, adopting and scaling a new public infrastructure such as dPIDs is difficult in a vacuum. Fortunately, the community has done extraordinary work with ORCID and RoR as identity layers for scientists and their organizations. Because we can build on existing identity solutions, adopting dPIDs does not need to start in a vacuum: we can both re-use existing infrastructure and benefit from its pre-existing network effects.
Additionally, trustworthy provenance combined with the open nature of the network would enable journals, libraries, or data stewards to add new information to enrich research objects over time. These dPID data enrichments could vary widely in their nature: from FAIR metadata (e.g., ontologies and controlled vocabularies), open peer-review reports, digitally verified badges for open data, reproducibility, and much more. This has many benefits, including creating better metrics and incentives for open science practices. Furthermore, since reading and writing on this content addressable network is without specific charges, equitable access is ensured, and substantial cost savings can be realized.
Compatibility of dPIDs and DOIs
It’s important to note that dPID is a new PID technology, not a new PID standard. The distinction is important because a proliferation of PID standards is not desirable. In fact, DOIs can be “upgraded” to become dPIDs by adding a DOI as a synonym to a dPID. DOIs would then simply resolve to a resource linked to a dPID filesystem. This backward compatibility would not only get rid of the need to update DOI records manually and make DOIs actually persistent, but it would also unlock the new capabilities described above. A single DOI could unfold into an entire file system with individual PIDs for machine-actionable digital objects and resolve deterministically to their mapped resource.
Beyond these properties, dPID technology can help to preserve the freedom and sovereignty of all stakeholders in the scientific ecosystem. Platform operators, repositories, publishers, and libraries maintain control over the content they share publicly. Importantly, dPID technology is entirely based on open-source software that anyone can run for free on their own hardware.
dPID has garnered much interest in the persistent identifier community. International Data Week 2023 saw the formation of a dPID Working Group with diverse participants from organizations around the globe. This group, hosted by the DeSci Foundation, aims to act as a driving force, community hub, and shared knowledge base and is open to anyone interested in participating.
Realizing the ideal of a fully machine-actionable scientific record is more critical than ever. While the importance of scientific data as a primary output of research is rising and data volume is increasing rapidly, interoperability needs to catch up. With the rise of AI and fake science, a trustworthy track record of a manuscript’s origin and context is invaluable. Given their properties, dPIDs and the deterministic resolution they provide are paths worth exploring.
The Tech Stack behind dPIDs
For those who would like to dive into the technical details, here is a brief overview of the core technologies that dPIDs are built upon. These protocols and software-based solutions have already been developed alongside their W3C formal specifications. They are free to use and open-source, enabling anyone to participate in further improvements.
- IPFS allows you to share and access data by content identifiers. Content hashes are used to identify and resolve each file. IPFS forms a peer-to-peer storage network of content-addressed information, allowing users to store, retrieve, and locate data based on a digital fingerprint of the actual content. This fingerprint is generated by a cryptographic hash function (e.g. SHA-256), which converts any content into a fixed-length string. Changing anything in the content (i.e. a single word, pixel, comma) will yield a different hash. SHA-256 allows generating 1077 different hashes, which is billions of times more than the number of atoms on Earth. Thus, the probability of two different inputs yielding the same hash is near zero. Because all data is guaranteed a unique fingerprint, content drift cannot happen because that would be accessible under a different fingerprint. Furthermore, it’s easy to check if the content you received from the IPFS network matches its hash, eliminating the risk of downloading content from unknown network peers. And thanks to IPLD, IPFS works not only with files but also with arbitrary data structures. This ensures the persistence of relations between artifacts such as manuscripts, data, code, or other PIDs for organizations and people. It also creates the possibility of mitigating the scientific record’s fragmentation and getting much better metadata and analytics.
- Decentralized Identifiers. Decentralized Identifiers (DIDs) enable verifiable, decentralized digital identity. A DID can refer to any target (e.g., a person, organization, thing, data model, abstract entity, etc.) as determined by the controller of the DID. The design enables the controller of a DID to prove control over it without requiring permission from any other party, allowing trustable interactions.
- Blockchain technology creates persistent recording. A blockchain is a distributed ledger with a growing list of records (blocks) securely linked via cryptographic hashes. Smart contracts associated with blockchains autonomously record the root hash of an IPLD folder structure on this cryptographic ledger, along with a time stamp and the ID of the person or entity who updated the record, creating highly persistent, open, and trustworthy metadata.
For those interested in the dPID Working Group mentioned above, newcomers are always welcome, and those interested can reach out to info@descifoundation.org.
Discussion
7 Thoughts on "Guest Post — Navigating the Drift: Persistence Challenges in the Digital Scientific Record and the Promise of dPIDs"
Sounds great. But I’m not quite sure what to think of it. Is it too good to be true or is it reinventing git (I think that uses MD5 hashes and commit history in a version controlled way). I can link to a particular version in a Github repository by citing a particular commit or file version. Where is the need to decentralise, go peer to peer and use blockchain? Why not just fork git, add some persistence features (eg disable deletion of public commits) and auto-deposit DOIs based on repository metadata. Github even has a long term preservation plan, so we know it can be archived in a persistent way.
Git doesn’t offer efficient data structures for sharing large files. The structure used in Git to store large files are called blobs (binary large objects) and blobs can easily become huge and unwieldy! If you’re required to download the whole blob for a large dataset you could exceed the limits of your disk, depending on the use case. IPFS on the other hand is built for efficient sharing of datasets because it uses *block* storage to split the large datasets into tiny chunks that can be spread across multiple disks while still being hash-verifiable to maintain data integrity. This allows you to efficiently share large files across a network, while being confident the data is correct. This is important when considering that not one institution, company, or entity may have all the hardware resources required to store all the datasets gathered by the scientific community. Additionally, with Git alone there is no concept of a verified timestamp. All timestamps are self-reported by users or rely on third party servers with fragmented and typically undocumented data-management policies. With blockchain, timestamps are derived via the consensus mechanism of the network, and are typically a transparent and documented feature of the protocol, solving the so-called “chronological oracle” problem (https://identity.foundation/sidetree/spec/#introduction) Knowing exactly when something was published can be crucial when dealing with issues like proper crediting of work and properly tracing retracted or edited publications, basically making it much easier to audit the entire trace of research activity, while having the advantage of creating a mechanism to discover potential unethical behavior or collusion — in a way that’s guaranteed, as opposed to the current status quo.
When we use Git on a server, we actually use a synthetic RPC to call some software via the network instead of accessing the data and treating them in compliance with the edge computing principle. The great example here is the difference between the data storage schemas in Git, as we can see from the repositories maintained by the IPFS bridge program I initially developed (https://github.com/ElettraSciComp/Git-IPFS-Remote-Bridge). This is an obvious 3-stated paradigm to keep all the branches and versions maintained properly. But for being accessible remotely from the same Git software, all the data and metadata should be converted into a special format that another instance of Git may understand too! Looking at the possibility of storing this specially formatted metadata and the actual data together in IPFS, we again see only the storage model instead of the server-side treatment model. And here the points of what we really need to store and treat versioned data in the network:
– Storage layer (IPFS + IPLD)
– Metadata + storage schema (in the case of Git – the baremetal repository format)
– Access model (for Git not yet implemented locally)
Thinking about all these things, I can easily imagine the data object representing something like edge-computing local Gitlab that does not need any server to run, and it would store and communicate the encrypted data together with corresponding metadata on IPFS (as just the separate data objects), supporting the access model controlled by the user-side of 3rd-party side trusted PKI. So, the point here is a question: “Why do we really need a server here, for this task?”. I think, this question should be revised for all the Internet, and it should be rethought for the cases where the server’s function is actually only to store the data and communicate the access controlled by someone else.
Adding to the replies above: Git functionality was an inspiration for this PID technology. That being said, Github’s implementation of git works flawlessly because one company with one backend stores and manages all of the data. This is undesirable because it creates data silos that can be exploited, as well as a single point of failure (i.e. the company controlling the data silo). P2P networks eliminate this problem. They also eliminate the need for a complex and expensive web of APIs connecting organizations, helping us facilitate more efficient collaboration across organizational boundaries, enabling data sovereignty, and creating the possibility for several organizations to store the same data using the same identifier (i.e. many copies keep things safe).
Great post. From Latin America with are working in a very similar idea, a Blockchain based implementation of ARK PIDS. (and IPFS for metadata layer) Our main goal is to build the basis for a public good PID service for our research institutions.
https://doi.org/10.5281/zenodo.8091668
I will contact the group in order to join efforts, I think that we have a lot in common.
Dear Christopher, Philipp, and Erik,
With interest I read your guest post in the Scholarly Kitchen
“Persistence Challenges in the Digital Scientific Record and the Promise of dPIDs” published on 2024-03-14.
I was encouraged by your overview of the current issues with reliably referencing digital scientific content: we do have a growing body of evidence to support that claim that original digital scientific works are lost due to link rot and content drift.
However, I was a bit surprised by your description of the dPID and their underlying technology Interplenatary File System (IPFS), especially following the meetings and email exchanges we’ve had in June/July of 2023 in context of NanoSessions #5/#6 https://nanopub.net/sessions .
In your 2024-03-14 blog post, you mention:
“[…] The core idea is that PIDs should not ask, “What is the content stored at this location?” Instead, the right question is, “What is the content with this digital fingerprint?” […]”
Then, a little later, you say:
“[…] IPFS […] allowing users to store, retrieve, and locate data based on a digital fingerprint of the actual content. This fingerprint is generated by a cryptographic hash function (e.g. SHA-256), which converts any content into a fixed-length string. […]”
However, as far as I know, IPFS does *not* allow for retrieval of content by their digital fingerprint in the form of a sha256 hash. Instead, a complex, hard to reproduce, CID (IPFS’s concept for a content identifier) is needed to retrieve specific content.
Following our 2023 exchanges, I tried to answer the question:
How can I retrieve a file from IPFS using their sha256 fingerprint?
And after several attempts, I keep getting stuck on the complexities of calculating CIDs (I think you call them dPIDs). You can see some of my attempts at https://github.com/bio-guoda/preston/issues/253#issue-1799634040 .
I think it would be nice to be able to building a bridge from IPFS infrastructures to the non-IPFS systems. But, as Ben Trask said in his 2015 exchange with Juan Benet (creator of IPFS) in https://github.com/ipfs/kubo/issues/1953, IPFS is block centric, not file-centric, which makes asking for file content rather complicated, and integration difficult.
In other words, I’d like to challenge your statement:
“[…] it’s easy to check if the content you received from the IPFS network matches its hash […]”
because I have not been able to do so independently.
And, this makes me wonder, why use IPFS CIDs to refer to content? Why not just use plain hashes (e.g., md5, sha256) of associated (meta)data as described in Elliott et al. 2023. (disclaimer: I am a co-author)?
Looking forward to your response,
-jorrit
https://jhpoelen.nl
https://linker.bio
# References
Elliott M.J., Poelen, J.H. & Fortes, J.A.B. (2023) Signing data citations enables data verification and citation persistence. Sci Data. https://doi.org/10.1038/s41597-023-02230-y https://linker.bio/hash://sha256/f849c870565f608899f183ca261365dce9c9f1c5441b1c779e0db49df9c2a19d
As we know from the IPFS CID spec, CID is a prefix tree representing actually a root node of the DAG. DAG here is the necessary element to ensure the data integrity on the binary (bitstream) level and distribute them on the network. So, IPFS operates over the chunked data gathered in the DAG, and the request returns block, but not files. This is a cost we should pay to obtain the possibility of retrieving the data from separate untrusted hosts. Thus, to recreate the CID locally, we should also recreate the chunking algorithm for the given bitstream, as explained here:
https://stackoverflow.com/questions/76799279/compute-cid-of-data-in-go
it is possible, and my example in Go there shows the way. The following Github repository contains very simple example program to recreate a dummy DAG with current IPFS defaults and to calculate proper CID in a standalone mode: https://github.com/twdragon/ipfs-cid-local However, it does not support calculations recursively over directories, as it requires additional efforts that I cannot dedicate right now to it.
Anyhow, the direct location of the file in the IPFS network via its OS-native SHA256 hash is not possible, except in some special cases, because CID by default represents a root DAG node associated with a file and its metadata.
However, there is an obvious implementation cost that exists here. We cannot use the file-based addressing over the SHA256 hashes because otherwise, we would lose the distributed nature of IPFS with its automated redundancy. To implement this kind of addressing, we have the following opportunities:
– Introduce SHA256 hash on the file level as an indexed default field of the lingered metadata, so it could be used as an input key for reverse lookup and resolution.
– Standardize the chunker routine used while addressing objects, and indicate it, so the CID recreation would be possible.
– Indicate the exact chunker routine used to store the objects together with their CIDs, in the default metadata fieldset.
– Borrow the versioned file-based storage model from VCS like Git. Obviously, it requires us to drop off IPFS and use the different custom storage layer.
Summarizing, your challenge over the sentence: “[…] it’s easy to check if the content you received from the IPFS network matches its hash […]”, – is completely understandable under the constraints of current IPFS implementation, but anyhow, it is possible to check the integrity of the DAG representing the file.
You raised also many very important questions about pure technical conditions we should set up for ourselves to obtain true persistence of the DAGs we store. They should be addressed and elaborated on as well.




